Data processing
Outbreak analyses are based on minimum spanning tree(MST)-visualizations of infection cases linked by genetic distances or contact tracing events. These MSTs rely on an imported dataset consisting of cases, sequenced samples, and contact information.
File imports
Case data
Cases are registered infection reports from health authorities. These are recorded using the SurvNet software developed by the RKI. Personal data is handled and stored exclusively on the client side. Addresses and names are used to create contact edges between cases, as it is valuable information if cases live at the same address (shared apartments, retirement homes, etc.) or have the same last name (potential family members).
A CSV structure with relevant fields was created for importing case data from SurvNet:
| Field | Naming options | Description | Required |
|---|---|---|---|
| Case id | Fall ID, Aktenzeichen | Unique ID of the case | ✅ |
| Registration date | Registrierungsdatum, Meldedatum | Date of registration in SurvNets | ✅ |
| Sequence id | Sequenz ID | Fasta ID of the corresponding sequence | |
| Outbreak | Ausbruch, AusbruchInfo_InternalName, AusbruchInfo_NameGA, AusbruchInfo_NameLS, AusbruchInfo_NameRKI,AusbruchInfo_GuidRecord, AusbruchInfo_Aktenzeichen, AusbruchInfo_InterneRef | Suspected outbreak association | |
| Infected by | Angesteckt bei, AngestecktBei | Unique ID of a case that was given as the origin of the infection | |
| Firstname | Vorname, PersonVorname | First name of the person associated with the case | |
| Lastname | Nachname, PersonFamilienname | Last name of the person associated with the case | |
| City | Ort, PersonOrt | Place of residence of the person associated with the case | |
| Zip code | PLZ, PersonPLZ | Zip code of the person associated with the case | |
| Street | Straße, PersonStrasse | Street of the person associated with the case | |
| Flexible category | Kategorie:{category_name} | Flexible category for further differentiation |
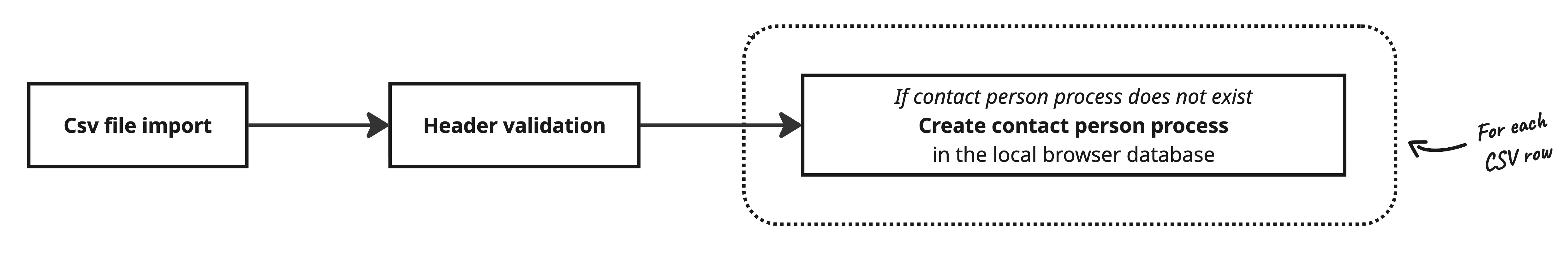
Case processing
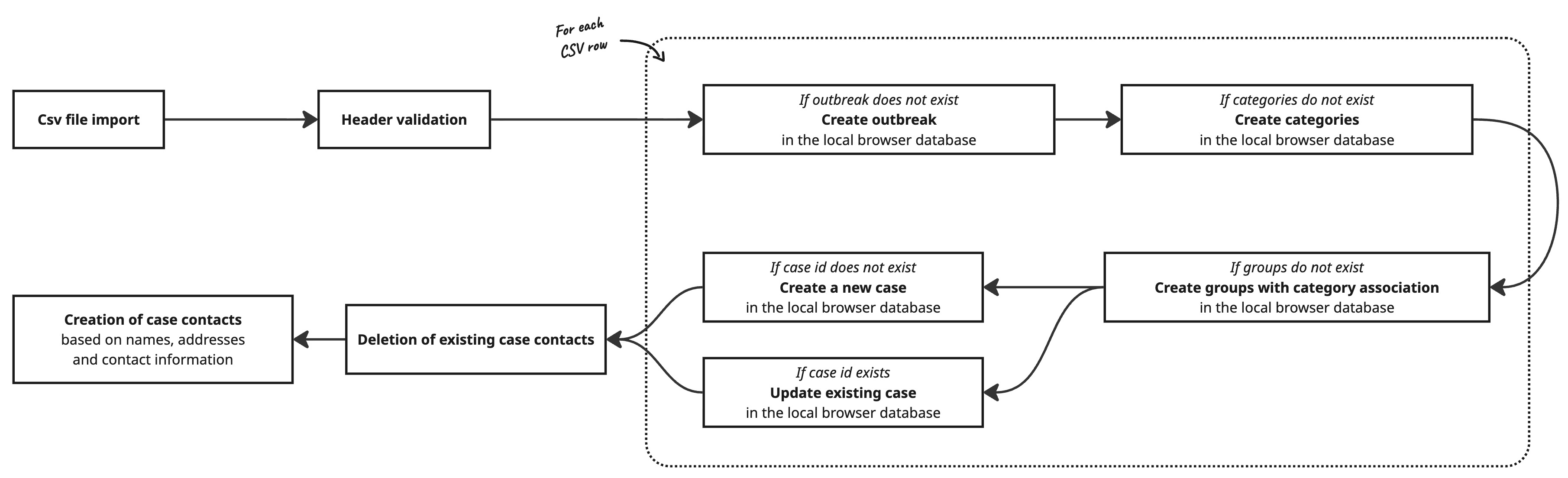
Sequence Data
Genetic distances are calculated between all imported sequences. To do this, a compressed representation of the sequences is first generated based on the present mutations. In the case of viruses, GENTRAIN uses Nextclade, while chewBBACA is used to determine existing alleles.
Based on the mutations, genetic distances between genome sequences can then be calculated and summarized in a distance matrix. This distance matrix enables the creation of a minimum spanning tree for the cases based on genetic distance.
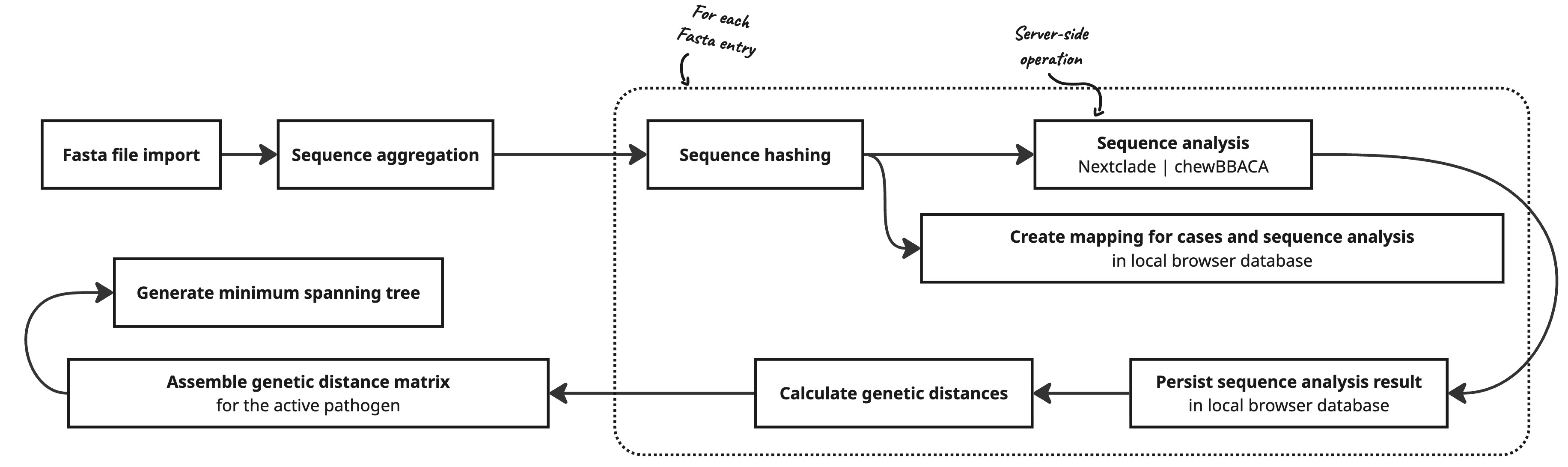
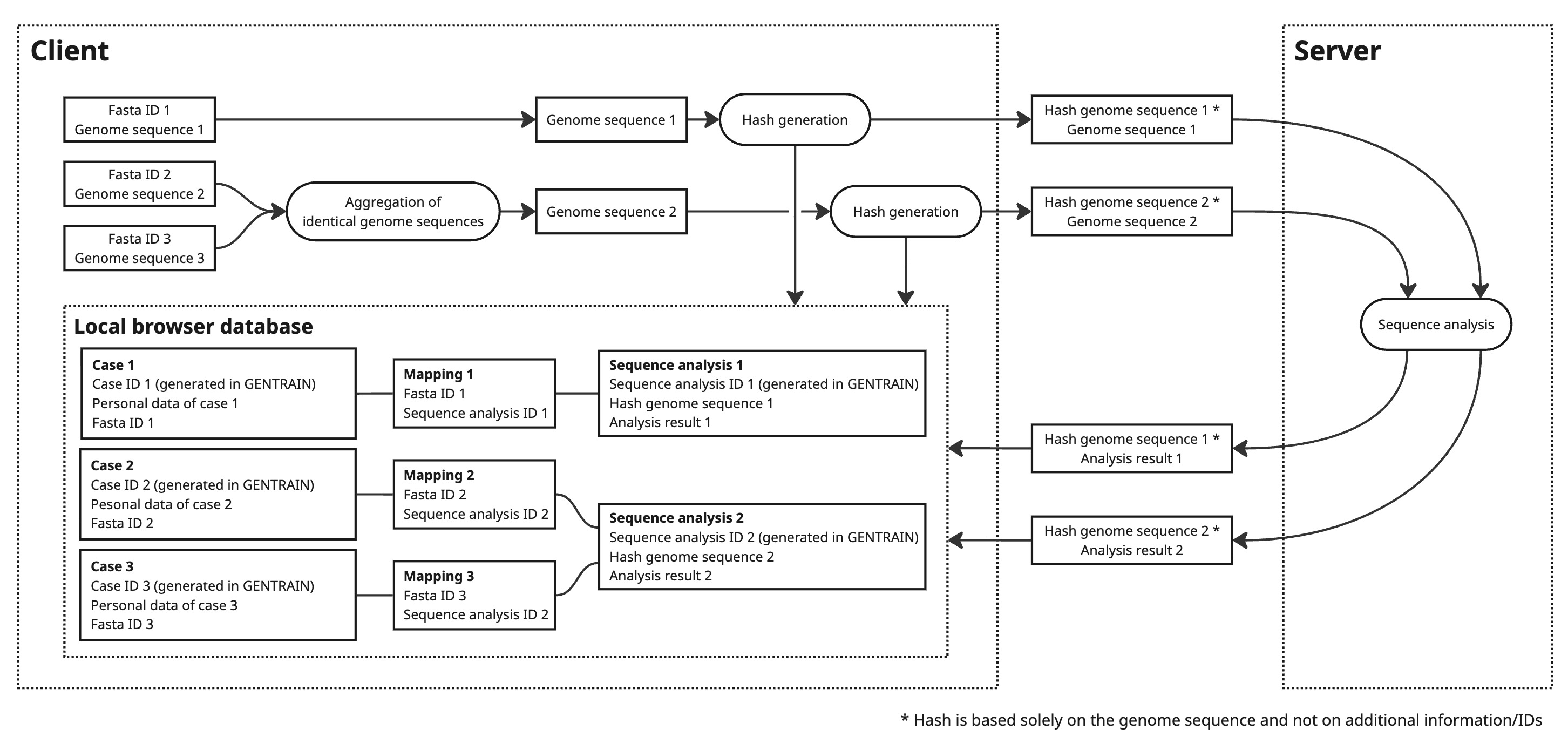
Aggregation of Identical Sequences
Cases may have identical genome sequences. To minimize storage space and ensure server-side anonymization, identical sequence data is aggregated into a single sequence analysis in GENTRAIN. For client-side mapping between cases and sequence analysis results, hashed sequences are used.
WebSocket Communication
Since sequence analysis can take several minutes, it is processed as jobs in a Redis queue. Therefore, a persistent channel via WebSockets was established for communication between client and server to ensure reliable information exchange. The WebSocket server is implemented using Flask SocketIO, and WebSocket clients using Socket.io.
- Creation of a WebSocket room for the analysis process
- Chunking of sequence data
- Transmission of sequence data chunks (with sequence hashes for identification) to the WebSocket room
- Once all chunks have been transmitted: enqueueing of the sequence analysis job in the Redis queue
- After completion of the sequence analysis job: transmission of the sequence analysis result to the WebSocket client
Sequence Analysis
To calculate genetic distances between viral and bacterial genomes, we analyze genetic sequences based on their reference genome. This allows us to maintain a minimized data structure without significant loss of information. Since viral sequences are typically fully available, and bacterial sequences are usually assembled, different analysis techniques are applied.
Viral Sequence Analysis
For viral sequences, mutations are determined based on the corresponding reference genome using the Nextclade CLI. Nextclade provides mutation objects consisting of SNPs, insertions, deletions, Ns, and non-ACGTN characters.
These mutation objects allow us to calculate genetic distances without storing the full sequence. All sequences in a fasta file are analyzed simultaneously as part of a single job. The fasta content is anonymized by replacing sequences with hashed sequences on both the client and server sides throughout the entire analysis process.
Nextclade provides a set of information for each analyzed sample. This information can be stored in various file formats; in our case, we receive a JSON object containing the detected clade, sequence quality metrics, and mutation data. Detected substitutions, insertions, deletions, missing characters, and non-ACGTNs are stored in the user's browser. This information allows us to reconstruct sequences — considering alignment — without needing the full sequence string.
{
substitutions: [
{pos: 209, refNuc: 'G', qryNuc: 'T'},
{pos: 240, refNuc: 'C', qryNuc: 'T'},
{pos: 3036, refNuc: 'C', qryNuc: 'T'},
...
{pos: 29741, refNuc: 'G', qryNuc: 'T'}
],
insertions: [
{pos: 18099, qryNuc: 'TCG'},
{pos: 29741, qryNuc: 'ACGT'}
],
deletions: [
{
range: {begin: 28247, end: 28253}
}
],
missings: [
{
character: 'N',
range: {begin: 0, end: 54}
},
{
character: 'N',
range: {begin: 6839, end: 6840}
}
],
nonACGTNs: [
{
character: 'Y',
range: {begin: 4504, end: 4505}
},
]
}
It may occur that a sequencing technology cannot unambiguously determine a nucleotide at a specific position. In this case, an ambiguous symbol is assigned that can represent all possible nucleotides at that position. If all four nucleotides are possible, for example, an “N” is used. These ambiguous symbols are defined in the IUPAC nomenclature.
{
"A": ["A"],
"C": ["C"],
"G": ["G"],
"T": ["T"],
"U": ["U"],
"M": ["A", "C"],
"R": ["A", "G"],
"S": ["C", "G"],
"W": ["A", "T"],
"Y": ["C", "T"],
"K": ["G", "T"],
"V": ["A", "C", "G"],
"H": ["A", "C", "T"],
"D": ["A", "G", "T"],
"B": ["C", "G", "T"],
"N": ["A", "C", "G", "T"],
"X": ["A", "C", "G", "T"]
}
Bacterial Sequence Analysis
Bacterial sequences are analyzed using chewBACCA. According to its own documentation, “chewBBACA is a software suite for the creation and evaluation of schemas and results for core-genome and whole-genome MultiLocus Sequence Typing (cg/wgMLST).”
For Gentrain, we use the AlleleCall service of chewBACCA, which provides mappings between each reference gene and the corresponding allele in the sequences. Based on these mappings, we then calculate the genetic distances by comparing the allele sets of the two sequences.
Each bacterial sequence assembly is provided in a separate FASTA file. Therefore, the import of bacterial sequences allows multiple files to be uploaded simultaneously. The filenames must match the FASTA IDs associated with the uploaded case data. These FASTA IDs are pseudonymized before communication with the server and stored exclusively on the client side.
The bacterial sequence analysis results in a mapping between the reference gene and the allele sequence of the corresponding sample (gene ID: allele sequence). These allele sequences are also hashed to minimize sequence length and ensure schema independence.
{
SAUR0001: "d3627b0e335350fc61d130d50e6516b2",
SAUR0002: "34d94e7230113031e160b5880f0ed5af",
SAUR0003: "ddf68eaae82c14021c4f44f73ac0789f",
...
SAUR3016: "e25053695fa8a872d478c73aba78c26a"
}
Distance Calculation
Genetic distances can be calculated between all cases of the active pathogen for which a sequence analysis result is available. Depending on whether the pathogen is viral or bacterial, these distances are determined differently.
Viral Distance Calculation
Genetic distances are calculated pairwise. Based on the Nextclade analysis results and the reference genome, complete sequences are reconstructed. Depending on the mutation found at a position of the reference genome, the following rules are applied.
| Mutation Type | Rule for Sequence Reconstruction |
|---|---|
| Substitution | The substituting nucleotide is added to the sequence that shows a substitution at the current reference position. Ambiguous symbols from the IUPAC nomenclature (including Ns) are also treated as substitutions. |
| Insertion | The inserted nucleotides are appended to the sequences that show an insertion at the current reference position. If both sequences contain an insertion at this reference position, they are aligned to identify the overlap of both insertions. For this alignment, the PairwiseAligner from the Biopython package is used. This package uses the Smith-Waterman algorithm, a widely recognized method for determining optimal alignments in genome sequences. If a sequence does not contain an insertion at this position, the inserted positions are filled with gaps. |
| Deletion | A gap is added to any sequence that shows a deletion at the current reference position. If both sequences show a deletion, the reference position is skipped entirely and not added to the reconstructed sequences. |
| No Mutation | The reference nucleotide for the current position is added to the sequences that show no mutation at that position. |
Next, the symbols at each individual position of the reference genome are compared with the corresponding positions in the two reconstructed sequences. If different symbols are present at a reference position, the distance is incremented depending on the symbols — or not. The following rules apply.
| Symbol | Rule for Distance Calculation |
|---|---|
| Missing (N) | If one of the sequences shows an N at the current alignment position, the genetic distance is not increased to avoid false-negative results. |
| Other ambiguous symbols | If a sequence shows a gap at the current alignment position, the genetic distance is only increased if this sequence did not have a gap at the previous alignment position. |
| Gaps | The genetic distance is only increased if the symbol at the current alignment position cannot represent the nucleotide of the other sequence at that position according to the IUPAC nomenclature. |
Bacterial Distance Calculation
For bacterial samples, this process is straightforward, as only the allele hashes per gene need to be compared. Different allele hashes result in a distance increment of 1. The distance value is not increased if the allele of a sample could not be determined by chewBACCA, in order to avoid false-positive distance increments.
Contact person data
Contact person records provide information about which cases have been in contact with each other. This information expands the contact information we extract from addresses and names. Importing contact person data creates contact edges labeled as 'Contact person'.
| Field | Naming Options | Description | Required |
|---|---|---|---|
| Case ID 1 | Fall ID 1 | Unique ID of the first case in the contact-person record | ✅ |
| Case ID 2 | Fall ID 2 | Unique ID of the second case in the contact-person record | ✅ |